Information
Journal Policies
Correlation between Mutations at the Beginning of Treatment and Virological Failure after 6 Months of Antiretroviral Treatment in People Living with the Human Immunodeficiency Virus in Kinshasa
Erick N. Kamangu1,2*
2.Research Group "HIV/AIDS Focus", Kinshasa - Democratic Republic of Congo.
Copyright : © 2018 . This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
The first-line Antiretroviral Therapy (ART) failure rate for Kinshasa was 16% in 2012 and 24.6% in 2014. The treatment failures can be caused by the presence of mutations transmitted and acquired by the virus.
The objective of this studywas to determine the correlation between the different HIV mutations transmitted at the beginning of treatment and the risk of virological failure after 6 months of treatment in naïve patients in Kinshasa.
One hundred and fifty-three (153) ART-naive patients were selected from 8 Ambulatory Treatment Centers in Kinshasa. Viral Loads (VL) and different strains of HIV-1 were determined for all patients at baseline as well as the different mutations associated with ART resistance. After RNA extraction, Reverse Transcription PCR (RT-PCR) and Nested PCR were performed to amplify the regions of interest for Protease and Reverse Transcriptase (RT) for sequencing. These fragments were sequenced by the Sanger sequencing method. The pairing of the obtained fragments was done with the Vector NTI Advance® 11.5 software and compared with different databases for the identification of HIV-1 subtypes and mutations.
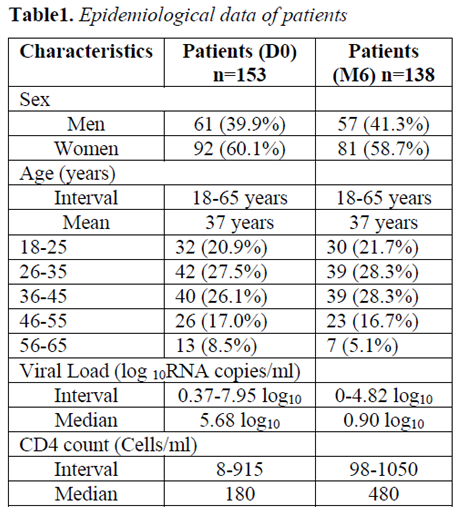
153 naïve ART patients were selected for follow-up at baseline. By the 6th month, 138 patients (90.2%) had returned for control. Eighty-one (58.7%) patients were women. The dominant age groups were 26 to 35 years and 36 to 45 years with 39 patients (28.3%) each. The most observed major mutations associated with Protease Inhibitors (PIs) were: L90 (2.0%), D30 (1.3%), V32 (1.3%), V82 (1.3%) and I84 (1.3%). The most frequent mutations for Nucleoside Inhibitors of Reverse Transcriptase (INTR) were: V75 (18.3%), K70 (9.8%), D67 (9.2%), M184 (9.2%) and T215 (9.2%). The most common mutations for Non-Nucleotide Inhibitors (NNRTIs) were: V179 (9.8%), K103 (8.5%), V106 (7.2%), Y181 (5.8%) and V90 (5.8%). At month 6, 104 patients (75.4%) had a VL less than 2.3 log10 RNA copies/ml giving a virological failure rate of 24.6%. According to the Pearson test, the failures of ART in the 6thmonth were strongly correlated with the K70 and V75 mutations for the INTRs as well as the V108 mutation for the INNTRs.
Mutations on K70 and V75 codons for Nucleotide Inhibitors as well as V108 for non-nucleotides are strongly implicated for first-line treatment failures in our population.
D0 mutations, antiretroviral therapy, HIV, PLHIV, Kinshasa
1. Introduction
The Human Immunodeficiency Virus (HIV) is the virus that causes the Acquired Immune Deficiency Syndrome (AIDS). Its genetic diversity influences the treatment and the emergence of resistance to treatment [1]. Some subtypes may develop resistance more frequently than subtypes A and B; this could be caused by some natural nucleotide polymorphisms on specific codons [1].
The prevalence of HIV infection in Kinshasa was reported to 1.6% according to the Demographic Health Survey (DHS) for 2013-2014 [2]. The first-line antiretroviral therapy (ART) failure rate for Kinshasa was of 16% in 2012 [3] and of 24.6% in 2014 [4]. Therapeutic failure brings together a wide range of situations, from virological failure resulting from persistent viral replication 6 months after treatment (Viral load> 200 RNA copies/ml or 2.30 log10 RNA copies/ml), immunological failure with persistence of immune deficiency (CD4 count < 200 cells/mm3) or clinical failure that usually associates virologic failure with immune damage [5]. These failures of ART can be caused by non-adherence and poor adherence to ART, the poor quality of the drugs dispensed the diversity of the virus as well as the presence of transmitted and acquired mutations [4].
The objective of this study was to determine the correlation between the different mutations transmitted at the beginning of treatment and the risk of virological failure after 6 months of treatment in naive patients in Kinshasa.
2. Methods
One hundred and fifty-three (153) antiretroviral treatment-naive patients (ART) were selected from 8 Ambulatory Treatment Centers (ATC) in Kinshasa. The inclusion criteria were: (i) be diagnosed HIV-1 positive according to national guidelines [6], (ii) aged of 18 years or older by date of inclusion in the cohort, (iii) eligible for ART in the monitoring ATC and (iv) naive of ART. Viral Loads (VL) and different strains of HIV-1 were determined for all patients at baseline and the different mutations associated with ART resistance [7]. At the 6thmonth of ART, only 138 patients (90.2%) of the cohort were received out of the 153 included on the 1stday of the study.
Clinical and paraclinical parameters were collected from individual patient survey cards. At the inclusion and at the 6thmonth, the determination of the VL was made at the laboratory of Molecular Biology of the Faculty of Medicine of the University of Kinshasa (UNIKIN) using an “in-house” assay previously described [8,9].
RNA was extracted from 140µl of plasma using the QIAamp RNA Mini Kit from QIAGEN® [10]. The extracted samples were stored at -80°C until use.
After extraction, a Reverse Transcription PCR (RT-PCR) followed by a Nested PCR were performed to amplify the regions of interest for Protease and Reverse Transcriptase (RT) for sequencing [11]. The fragments were sequenced by the Sanger sequencing method, purified by the ExoSap-IT technique to remove residual PCR products.
The pairing of the obtained fragments was done with the Vector NTI Advance® 11.5 software (Invitrogen, Life Technologies) and compared with different databases (Stanford University, ANRS and Max Plank) for the identification of the HIV-1 subtypes and different mutations.
The clinical and laboratory parameters of the 6thmonth were compared with those of the inclusion of the cohort in order to determine the evolution of the patients under treatment. For compliance, the clinical profiles were evaluated according to the World Health Organization (WHO) classification; the VLs were determined under the same conditions. Correlation tests were done between VLs at baseline and those at the 6thmonth, VLs at inclusion and mortality, VLs at baseline and treatment failure, as well as prevalence of acquired mutations and treatment failures.
The Pearson correlation test was used to analyze the data. The value of p < 0.05 was considered significant.
Virological treatment failure was defined as a persistence VL greater than 200 RNA copies/ml (2.30 log10 RNA copies/ml) 6 months after the start of treatment [5]. Lost patients are patients who did not return for their 6th month appointment and who were not found by the community relay services of the respective centers.
3. Results
At inclusion, 153 antiretroviral treatment-naive patients were selected for follow-up. Ninety and one (60.1%) patients were women and 62 (39.9%) were men. The ages of patients were within the range of 18 to 65 years with a median of 37 years. The most represented age group was 26 to 35 years with 42 patients (27.45%) followed by those aged 36 to 45 years (26.14%). At the 6thmonth, 138 patients (90.2%) out of the 153 patients selected for inclusion had returned for the control. Eighty-one (58.7%) patients were women and 57 (41.3%) were men. The dominant age groups were 26 to 35 years and 36 to 45 years, with 39 patients (28.3%) each, followed by those aged 18 to 25 years (21.7%). Ten (10) cases of death, or 6.5%, were reported and 5 patients (3.3%) are lost from the community relays of the respective centers (Table 1).
Several mutations associated with Protease Inhibitors (PIs) have been observed. The major mutations of the most observed PIs were: L90 (2.0%), D30 (1.3%), V32 (1.3%), V82 (1.3%) and I84 (1.3%). The minor mutations in the most observed PIs were: K20 (28.1%), L10 (27.5%), I47 (6.5%), V11 (5.2%), V32 (4.6%), G48 (3.3%) and A71 (2.0%).
Some mutations associated with Nucleoside Inhibitors of Reverse Transcriptase (NRTI) and Non-Nucleotide Inhibitors of Reverse Transcriptase (NNRTI) have also been found. For NRTIs, the most common mutations were: V75 (18.3%), K70 (9.8%), D67 (9.2%), M184 (9.2%), T215 (9.2%), Y115 (7.8%), M41 (7.2%), T69 (5.2%), and L74 (3.9%). For NNRTIs, the most common mutations were: V179 (9.8%), K103 (8.5%), V106 (7.2%), Y181 (5.8%), V90 (5.8%), A98 (5.2%), V108 (5.2%), Y188 (4.6%) and F227 (4.6%).
At baseline, the median value of Viral Loads (VL) for enrolled patients was 5.68 log10 RNA copies/ml. The minimum and maximum values were respectively 0.37 log10 and 7.95 log10 RNA copies/ml with 97 patients (63.4%) having a VL greater than 100,000 RNA copies/ml or 5.0 log10RNA copies/ml.
At month 6, 5 patients were lost to follow-up (2 women and 3 men) and 10 patients died (8 women and 2 men) (Table 1). The median VL value of the patients was 0.90 log10 RNA copies/ml. The minimum and maximum values were respectively 0 and 4.82 log10 RNA copies/ml with 104 patients (75.4%) having a VL less than 200 RNA copies/ml or 2.3 log10 RNA copies/ml thus giving a virological failure rate of 24.6%. Of the 34 patients with virological failure, 8 (23.5%) had minimal failure (2.30 log10 < VL < 3.70 log10 RNA copies/ml), 23 (67.7%) had moderate failure (3.70 log10, < VL < 4.48 log10 RNA copies/ml) and 3 (8.8%) in severe failure (VL > 4.48 log10 RNA copies/ml).
According to the Pearson test, VLs at month 6 were strongly correlated with that of inclusion (R² = 0.641, p < 0.000), at the K70 mutation for NRTIs (R² = 0.558, p < 0.000) at the V75 mutation for NRTIs (R² = 0.448, p< 0.000), the V108 mutation for NNRTIs (R² = 0.413, p < 0.000), and virological treatment failure (R² = 0.947, p< 0.000).
4. Discussions
The objective of this study was to determine the correlation between the different mutations transmitted at the beginning of treatment and the risks of virological failure after 6 months of treatment in patients in Kinshasa.
At the start of treatment, the population consisted of 153 antiretroviral treatment-naive patients. One hundred and thirty (130) samples, or 84.9%, were amplified on the protease region and 145 samples (94.8%) on the Reverse Transcriptase (RT) region. This difference in amplification has also been previously described; samples are more easily amplified on TR than on Protease because of the size of this gene of interest [12, 13]. Subtype A was dominant at 22.9% of the study population. It was followed by CRF02_AG (11.1%), C (9.8%), G (9.8%), K (9.8%), D (7.8%), H (7.8%) and J (5.0%). Several primary resistances have been observed for PIs; major and minor mutations. The most important major mutations were: L90 (2.0%), D30 (1.3%), V32 (1.3%), V82 (1.3%) and I84 (1.3%). The most observed minor mutations were: K20 (28.1%), L10 (27.5%), I47 (6.5%), V11 (5.2%), V32 (4.6%), G48 (3.3%) and A71 (2.0%). The most common NRTI-associated transmitted mutations in this study population were: V75 (18.3%), K70 (9.8%), D67 (9.8%), M184 (9.2%), T215 (9.2%), Y115 (7.8%), M41 (7.2%), T69 (5.2%) and L74 (3.9%). For NNRTIs, there were: V179 (9.8%), K103 (8.5%), V106 (7.2%), Y181 (5.8%), V90 (5.8%), A98 (5.2%), V108 (5.2%), Y188 (4.6%) and F227 (4, 6%). For NRTIs, the T69 mutation (5.2%) is associated with resistance to all NRTIs; the K65 mutation (2.6%) is associated with resistance for Stavudine (d4T), Abacavir (ABC), Lamivudine (3TC), Tenofovir (TDF) and Didanosine (ddI); K70 (9.8%) is associated with TDF; L74 (3.9%) at ddI and ABC; V75 (18.3%) at d4T; the Y115 (7.8%) at ABC; M184 (9.2%) at 3TC; and T215 (9.2%) with Zidovudine (ZDV) and d4T [15,16].For NNRTIs, the A98 mutation (5.2%) is associated with resistance against Nevirapine (NVP) while E138 (3.3%) is implicated for Efavirenz (EFV); L100 (2.6%), K103 (8.5%), V106 (7.2%), Y181 (5.8%), Y188 (4.6%), G190 (3.9%) and M230 (1.3%) are all associated with NVP and EFV [15,17]. Hence, more than 10% of naïve patients in this population have a predisposition to first-line treatment failure as recommended by the National Program for the Democratic Republic of Congo (ZDV + 3TC + NVP) because of mutations transmitted during infection.
After 6 months of ART, 10 cases (6.5%) of deaths and 5 cases (3.3%) of lost-sighted patients were recorded in the cohort of patients followed. For the cases of death, the majority (8 patients) are women whereas for the lost-sighted they are men (3 men). Of the 138 patients (90.2%) who returned for their control, 81 (58.7%) patients were women and 57 (41.3%) were men (p < 0.06). Various other studies have also published similar ratios that point out that women are more affected than men by HIV infection in our environment [2,14,18].
The median CV value of the included patients was 0.90 log10 RNA copies/ml. The minimum and maximum values were respectively 0 and 4.82 log10 RNA copies/ml with 104 patients (75.4%) having a VL less than 200 RNA copies/ml or 2.3 log10 RNA copies/ml thus giving a virological failure rate of 24.6%. Of the 34 patients with virological failure, 8 (23.5%) had minimal failure (2.30 log10 < VL< 3.70 log10 RNA copies/ml), 23 (67.7%) had moderate failure (3.70 log10 < VL < 4.48 log10 RNA copies/ml) and 3 (8.8%) in severe failure (VL > 4.48 log10 RNA copies/ml). Most patients with treatment failure (67.7%) have moderate virological failure. In the past, virological failure was estimated at 14.6% in 2011 [19] and 16% in 2012 [3] for Kinshasa, taking into account 3 clinics of a reference structure of that time. The difference in numbers lies in the criteria for inclusion of patients and selection of centers, as well as in the scale of determination of treatment failure. Indeed, treatment failure was redefined as a VL > 200 RNA copies/ml (2.3 log10 RNA copies/ml) in 2013 [20] as opposed to a VL > 1000 RNA copies/ml (3.0 log10 RNA copies/ml) from previous years [5]. In the present study, were randomly selected 2 centers by district of Kinshasa that meet the criteria according to WHO recommendations [21]. Thus for Kinshasa, according to the updated criteria, the virological failure rate is estimated at 24.6%.
According to the Pearson test, VLs at month 6 were strongly correlated with that of inclusion (R² = 0.641, p = 3.25 X 10-4), with the K70 mutation for NRTIs (R² = 0.558; p = 4.76 X 10-4), the V75 mutation for NRTIs (R² = 0.448, p = 9.1 X 10-4), and the V108 mutation for NNRTIs (R² = 0.413; p = 7.6 x 10-4) and virological treatment failure (R² = 0.947, p = 8.3 X 10-5). Various studies have shown that high VL (VL > 5.00 log10) before treatment is a poor prognosis for treatment [22,23]. Some studies have incriminated K103N and Y181C mutations for treatment resistance and failure [24,25]. However in this case, the mutated codons that are responsible for treatment failure in naïve patients are: K70 NRTI, V75 NRTI and V108 NNRTI at the start of treatment. This presents a profile of the different resistance mutations for the city of Kinshasa specific to the different variants that correspond to what has been published in Africa for non-B subtypes [26].
5. Conclusion
These results confirmed the hypothesis that a high viral load at the beginning of treatment is a poor prognosis for the evolution of the patient. Mutations on the K70 and V75 codons for Nucleotide Inhibitors as well as V108 for non-nucleotides are strongly implicated for treatment failures in our population. Significant associations between virological failure, acquired mutations, and viral load at baseline reinforce the importance of the usefulness of genotyping and viral load testing at the beginning of treatment to improve treatment as well as adequate therapy.
References
- Shao Y, Williamson C. The HIV-1 Epidemic: Low- to Middle-Income Countries. Cold Spring HarbPerspect Med. 2012; 2: a007187.
- Ministère du Plan et Suivi de la Mise en œuvre de la Révolution de la Modernité, Ministère de la Santé Publique, République Démocratique du Congo (RDC). Enquête Démographique de la Santé (EDS-RDC). 2014
- Kamangu NE, Kawila ME, Mukumbi H, Mvumbi LG. Estimated rates of treatment failure in first-line antiretroviral treatment in Kinshasa: Case of the ACS AMO-Congo. International Journal of Collaborative Research on Internal Medicine and Public Health (IJCRIMPH). June 2012; 4 (6): 885-91
- Kamangu NE. Estimation of Clinical, Immunological and Virological Failure of First Line Antiretroviral Treatment in Kinshasa, Democratic Republic of Congo. Open Access Library Journal. 2018, 5: e4560. https://doi.org/10.4236/oalib.1104560.
- Yéni P. (sous la direction de). Prise en charge médicale des personnes infectées par le VIH. Recommandations du groupe d’Experts. Paris, France. Médecine Sciences, Flammarion. 2010 : 418.
- Programme National de la Lutte contre le Sida et les IST (PNLS), Ministère de la Santé Publique, République Démocratique du Congo.Guide National de Traitement de l’infection à VIH par les antirétroviraux chez l’adolescent et l’adulte ; Révision 2013.
- Kamangu NE, Bulanda IB, Bongenia IB, Botomwito TH, Mvumbi LG, De Mol P, Vaira D, Hayette MP, Kalala LR. Virological Profile of Patients Infected with HIV Starting Antiretroviral Treatment in Kinshasa. Open Access Library Journal. 2015, 2: e1564. http://dx.doi.org/10.4236/oalib.1101564
- Kamangu NE, Chatté A, Boreux R, Kalala LR, Mvumbi LG, De Mol P, Vaira D, Hayette MP. Implementation of an In-House Quantitative Real-Time PCR for Determination of HIV Viral Load in Kinshasa. Open Access Library Journal. 2014; 1: e855. http://dx.doi.org/ 10.42 36/oalib.1100855
- Kamangu NE, Chatté A, Boreux R, Susin F, Kalala LR, Mvumbi LG, De Mol P, Hayette MP, Vaira D. Comparison of an In-House Quantitative PCR and COBAS Ampliprep/ TaqMan Roche for Determination of Viral Load for HIV Type 1 non-B. Open Access Library Journal. 2015, 2: e1402. http:// dx.doi. org/10.4236/oalib.1101402
- QIAGEN. QIAamp® RNA Mini and Blood Mini Handbook. 3rd Edition. April 2010
- Steegen K, Demecheleer E, De Cabooter N, Nges D, Temmerman M, Ndumbe P, Mandaliya K, Plum J, Verhofstede C. A Sensitive in-house RT-PCR Genotyping System for Combined Detection of Plasma HIV-1 and Assessment of Drug Resistance. J Virol Methods. 2006; 133: 147-55
- Jordan MR, Winsett J, Tiro A, Bau V, Berbara RS, Rowley C, Bellosillo N, Wanke C, Coakley EP. HIV Drug Resistance Profiles and Clinical Outcomes in Patients with Viremia Maintained at Very Low Levels. WJA. 2013; 3: 71-8.
- Loukou YG, Zinzendorf NY, Kouadio H, Dje L, Cablan MA, Lathro SJ, Akoua Koffi MC. Genetic Diversity and Antiretroviral Drug Resistance among Drug-Naïve HIV-1 Infected Pregnant Women Attending Antenatal Clinics in Abidjan, Côte d’Ivoire. WJA. 2012; 2: 57-63
- Kamangu NE, Chatte A, Susin F, Boreux R, Kalala LR, Mvumbi LG, De Mol P, Vaira D, Hayette MP. Genetic Diversity and Antiretroviral Drug Resistance among Drug-Naïve HIV Type 1 Infected Patients attending Clinics in Kinshasa, Democratic Republic of Congo. Journal of HIV and AIDS. 2015; 1:1 http://dx.doi.org/10.16966/jha.101
- Margot NA, Lu B, Cheng A, Miller MD; Study 903 Team. Resistance development over 144 weeks in treatment-naive patients receiving Tenofovir Disoproxil Fumarate or Stavudine with Lamivudine and Efavirenz in Study 903. HIV Med. 2006; 7(7): 442-50
- Kagan RM, Lee TS, Ross L, Lloyd RM Jr, Lewinski MA, Potts SJ. Molecular basis of antagonism between K70E and K65R Tenofovir-associated mutations in HIV-1 Reverse Transcriptase. Antiviral Res. 2007; 75(3): 210-8
- Harrigan PR, Mo T, Wynhoven B, Hirsch J, Brumme Z, McKenna P, Pattery T, Vingerhoets J, Bacheler LT. Rare mutations at codon 103 of HIV-1 reverse transcriptase can confer resistance to non-nucleoside reverse transcriptase inhibitors. AIDS. 2005; 19 (6): 549-54
- Programme National de Lutte contre le VIH/SIDA et les Infections Sexuellement Transmissible (PNLS), Ministère de la Santé Publique, République Démocratique du Congo (RDC). Guide National de Prise en Charge de l’Infection à VIH en RDC. Version révisé 2013
- Muwonga J, Edidi S, Butel C, Vidal N, Monleau M, Okenge A, Mandjo JL, Mukumbi H, Muyembe JJ, Mbayo F, Kayembe D, Delaporte E, Boillot F, Peeters M. Resistance to Antiretroviral Drugs in Treated and Drug-Naïve Patients in the Democratic Republic of Congo.Journal of Acquired Immune Deficient Syndrome. 2011; 57 (Supl 1): S27-S33
- Morlat P (sous la direction de). Prise en Charge Médicale des Personnes Vivant avec le VIH: Recommandations du Groupe d’experts. Sous l’égide du CNS et de l’ANRS. Rapports 2013.
- Kamangu NE, Kalala NH, Mesia KG. Profile of Antiretroviral Treatment Centers in Kinshasa, Democratic Republic of Congo [Poster 388]. In proceedings of the 1st International African Society of Laboratory Medicine (ASLM) Conference. 1-7 December 2012; Cape Town, South Africa. 2012: 377
- Egger M, May M, Chêne G. Prognosis of HIV-1-Infected Patients Starting Highly Active Therapy: A Collaborative Analysis of Prospective Studies. Lancet. 2002; 360 (9327): 119-29
- Phair JP, Mellors JW, Detels R, Margolick JB, Munoz A. Virologic and Immunologic Values Allowing Safe Deferral of Antiretroviral Therapy. AIDS. 2002; 16 (18): 2455-59
- Anejo-Okopi JA, Agbaji OO, Agaba PA, Ugoagwu PO, Were K, Onywera H, Owiti P, Isa SE, Otecko N, Okwori AEJ, Musa J, Oguche S, Sagay AS, Idoko JA, Nimzing L,Jatau ED, Olonitola OS. Human Immunodeficiency Virus Type 1 (HIV-1) genetic diversity and prevalence of antiretroviral drug resistane mutations in treatment-naïve adults in Jos, North Central Nigeria. Afr. J. Biotechnol. 2013; 12 (17): 2279-87
- Afonso JM, Bello G, Guimaraes ML, Sojka M, Morgado MG. HIV-1 Genetic Diversity and Treatment Drug Resistance Mutations among Patients from North, Central and South Regions of Angola. PLoS ONE. 2012; 7 (8): e42996. http://doi:10.1371/journal.pone.00042996
- Santos AF, Soares MA. HIV Genetic Diversity and Drug Resistance. Virus. 2010; 2: 503-31. http://doi:3390/v2020503